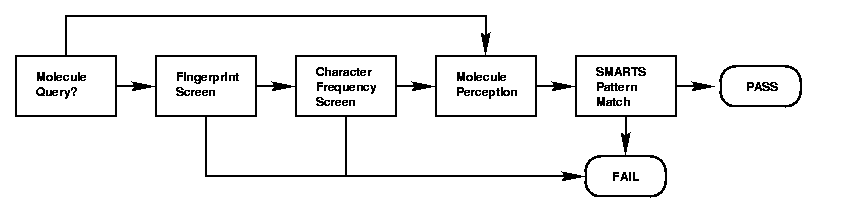
其大概æ„æ€å°±æ˜¯å…ˆé€šè¿‡Fingerprint˜q›è¡Œ½{›é€‰ï¼Œ˜q™æ ·å¯ä»¥å¿«é€Ÿçš„½{›é€‰æŽ‰ä¸€éƒ¨åˆ†æ•°æ®åQŒå¯¹äºŽå¤æ‚结构更有效åQ›å¦å¤–å°±æ˜¯æ ¹æ®åŽŸå个数或者特ŒDŠåŽŸå个数进行比较,如果查询¾l“构包å«ä¸‰ä¸ª“N”原ååQŒé‚£ä¹ˆæ‰€è¦æŸ¥è¯¢å‡ºçš„结构所å«æœ‰“N”的个数必™åÕd¤§äºŽç‰äº?åQŒè¿™æ ·å¯¹äºŽåŒ…å«ä¸€äº›ç‰¹ŒDŠå…ƒç´ 的效果是特别的好;˜q˜æœ‰ž®±æ˜¯æ ÒŽ®åˆ†å的一些性质˜q›è¡Œ½{›é€‰è¿‡æ»¤ï¼Œæ¯”如芳香性ç‰åQ›æœ€åŽå†˜q›è¡ŒåŒšw…åQŒè¿™æ ·ä¸€æ¥å¯¹äºŽå¤æ‚结构以åŠå«ç‰ÒŽ®Šå…ƒç´ 的查询速度会æ高很多ã€?br /> 最åŽæ–‡ç« 丘q˜ç»™å‡ºæµ‹è¯•æ•°æ®ï¼Œä»Žä¸å¯ä»¥çœ‹å‡ºåQŒé€Ÿåº¦ä¸€èˆ¬æ高了三å€å·¦å»I¼š
| Name | SMILES | Correct | FP | Triage | Before | After | Latest |
| Propane | CCC | 65337 | 66352 | 42411 | 42.59 | 17.99 | 14.34 |
| Selenium | [Se] | 246 | 995 | 225 | 0.80 | 0.83 | 0.52 |
| Benzene | c1ccccc1 | 79426 | 79486 | 50893 | 72.69 | 27.56 | 20.29 |
| Methane | C | 118519 | 118524 | 118511 | 61.29 | 5.47 | 4.25 |
| Amido | NC=O | 25695 | 26975 | 14702 | 18.89 | 9.84 | 8.16 |
| Methylbenzene | Cc1ccccc1 | 54529 | 56869 | 20490 | 54.76 | 35.58 | 25.90 |
| Carboxy | OC=O | 33009 | 34369 | 17809 | 23.86 | 12.48 | 10.24 |
| Chlorine | Cl | 19424 | 23318 | 19424 | 11.23 | 1.38 | 1.12 |
| Cyclopropane | C1CC1 | 863 | 4358 | 484 | 8.24 | 7.78 | 5.02 |
| Biphenyl | c1ccccc1c2ccccc2 | 2967 | 5142 | 146 | 21.94 | 21.65 | 11.44 |
| Dopamine | NCCc1ccc(O)c(O)c1 | 829 | 913 | 23 | 1.85 | 2.09 | 1.47 |
| Sulfisoxazole | 7 | 8 | 3 | 0.50 | 0.88 | 0.51 | |
| BetaCarotene | 2 | 16 | 1 | 0.48 | 0.68 | 0.58 | |
| Nitrofurantoin | 0 | 0 | 0 | 0.42 | 0.58 | 0.52 |
import Java.io.File;
import Java.io.FileNotFoundException;
import Java.io.FileReader;
import Java.util.List;
import org.openscience.cdk.ChemFile;
import org.openscience.cdk.ChemObject;
import org.openscience.cdk.Molecule;
import org.openscience.cdk.exception.CDKException;
import org.openscience.cdk.interfaces.IAtomContainer;
import org.openscience.cdk.io.MDLReader;
import org.openscience.cdk.io.MDLV2000Reader;
import org.openscience.cdk.tools.manipulator.ChemFileManipulator;
public class ReadSDFTest {
/**
* @param args
* @throws CDKException
* @throws FileNotFoundException
*/
public static void main(String[] args) throws CDKException, FileNotFoundException {
String filename = "H:\\molecules.sdf";
// InputStream ins = ReadSDFTest.class.getClassLoader().getResourceAsStream(filename);
// MDLReader reader = new MDLReader(ins);
//alternatively, you can specify a file directly
MDLV2000Reader reader = new MDLV2000Reader(new FileReader(new File(filename)));
ChemFile chemFile = (ChemFile)reader.read((ChemObject)new ChemFile());
List<IAtomContainer> containersList = ChemFileManipulator.getAllAtomContainers(chemFile);
Molecule molecule = null;
for (IAtomContainer mol : containersList) {
molecule = (Molecule) mol;
System.out.println(molecule.getProperties());
System.out.println(molecule.getProperty("CD_MOLWEIGHT"));
// Fingerprinter fp = new Fingerprinter();
// BitSet bt = fp.getFingerprint(molecule);
// System.out.println(bt);
}
}
}
Rich Apodaca wrote a great serious posts named Fast Substructure Search Using Open Source Tools providing details on substructure search with MySQL. But, however, poor binary data operation functions of MySQL limited the implementation of similar structure search which typically depends on the calculation of Tanimato coefficient. We are going to use Java & CDK to add this feature.
As default output of CDK fingerprint, java.util.BitSet with Serializable interface is perfect data format of fingerprint data storage. Java itself provides several collections such as ArrayList, LinkedList, Vector class in package Java.util. To provide web access to the search engine, thread unsafe ArrayList and LinkedList have to be kicked out. How about Vector? Once all the fingerprint data is well prepared, the collection function we need to do similarity search is just iteration. No add, no delete. So, a light weight array is enough.
Most of the molecule information is stored in MySQL database, so we are going to map fingerprint to corresponding row in data table. Here is the MolDFData class, we use a long variable to store corresponding primary key in data table.
public class MolDFData implements Serializable {
private long id;
private BitSet fingerprint;
public MolDFData(long id, BitSet fingerprint) {
this.id = id;
this.fingerprint = fingerprint;
}
public long getId() {
return id;
}
public void setId(long id) {
this.id = id;
}
public BitSet getFingerprint() {
return fingerprint;
}
public void setFingerprint(BitSet fingerprint) {
this.fingerprint = fingerprint;
}
}
This is how we storage our fingerprints.
private MolFPData[] arrayData;
No big deal with similarity search. Just calculate the Tanimoto coefficient, if it’s bigger than minimal similarity you set, add this one into result.
public List searchTanimoto(BitSet bt, float minSimlarity) {
List resultList = new LinkedList();
int i;
for (i = 0; i < arrayData.length; i++) {
MolDFData aListData = arrayData[i];
try {
float coefficient = Tanimoto.calculate(aListData.getFingerprint(), bt);
if (coefficient > minSimlarity) {
resultList.add(new SearchResultData(aListData.getId(), coefficient));
}
} catch (CDKException e) {
}
Collections.sort(resultList);
}
return resultList;
}
Pretty ugly code? Maybe. But it really works, at a acceptable speed.
Tests were done using the code blow on a macbook(Intel Core Due 1.83 GHz, 2G RAM).
long t3 = System.currentTimeMillis();
List<SearchResultData> listResult = se.searchTanimoto(bs, 0.8f);
long t4 = System.currentTimeMillis();
System.out.println("Thread: Search done in " + (t4 - t3) + " ms.");
In my database of 87364 commercial compounds, it takes 335 ms.
JChemPaint was started by Christoph Steinbeck in the late 1990's to be the complementary structure editor to Jmol. It was then co-developed by Egon Willighagen and others. Jmol again is a visualisation and analysis tool for 3D molecular structures, started by Dan Gezelter at Notre Dame University, initiator of the Open Science Project and, like JChemPaint, developed by an international team of opensource programmers.
In at least three aspects JChemPaint is different from other 2D editors:
- JChemPaint is open source and free software. We believe that scientific software, especially when its development was publicly funded, should be free. As the GNU people put it: «`Free software´ is a matter of liberty, not price. To understand the concept, you should think of `free speech´, not `free beer´». Everyone can participate in the development of the program. Everyone can download and change the source code, provided that they make the changes publicly available again, according to the GNU Lesser General Public License, LGPL. This ensures that the community can take advantage of any bugfix or enhancement made to the system. It also ensures that a scientist, who needs a standard piece of software like a structure editor as a helper application in his/her new program, does not have to reinvent the wheel over and over again because all the structure editors that have been written before are now proprietary software. If there is a free structure editor, he/she can focus on the real science.
- JChemPaint is in constant development and you can help (see below).
- Since JChemPaint is written in Java, it runs on any computing platform and operating system for which a Java Virtual Machine (of version >= 1.3 up to JCP 2.4 and version >= 1.5 for JCP > 2.4) has been implemented (like Linux, Windows, Solaris, AIX and others).
- JChemPaint is available free of charge.
- JChemPaint is translated into several languages: Dutch, French, German, Polish, Portuguese and Spanish.
<applet code="org.openscience.jchempaint.applet.JChemPaintEditorApplet"
archive="jchempaint-applet-core.jar" name="Editor"
width="550" height="400">
</applet>

集æˆViewerApplet到jsp™åµé¢é‡?
<applet code="org.openscience.jchempaint.applet.JChemPaintViewerApplet"
archive="jchempaint-applet-core.jar"
width="550" height="400">
Applet methods:
Reading from the applet
- getMolFile()
- getSmiles()
- getSmilesChiral()
- getParameter()
- getParameterInfo()
- getAppletInfo()
- getLocale()
- getImage()
- getTheJcpp()
Writing to the applet
- setMolFile()
- setMolFileWithReplace()
- addMolFileWithReplace()
- loadModelFromUrl()
- loadModelFromSmiles()
- clear()
- selectAtom()
- init()
- start()
- stop()
- initPanelAndModel()
- setTheJcpp()
- setTheModel()